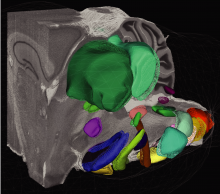
Construction of an active digital atlas of the mouse brain, with current emphasis on the brainstem. This is a full stack project that spans from data acquisition to imaging processing to computational anatomy. Involves Prof. Y. Freund in CSE, Prof. D. Kleinfeld in Physics and Neurobiology, and their professional associates.
Type of Data:
High resolution, light-level structural and functional images of the mouse brain
Approximate Data Size:
1 Tb per brain
Domain Expert:
Methods Expert:
Methods Student:
Methods Student Openings:
1.00
Methods Student Funding:
yes
Methods Student Prerequisites:
The main task is to write code that finds transformation of point sets in 3D. This requires a solid foundation in linear algebra and optimization.
Code is to be written in python / numpy. Code has to be well documented and easy to read. Working well with others is a must.
